Note
Go to the end to download the full example code
Ramachandran plot of dynein motor domain#
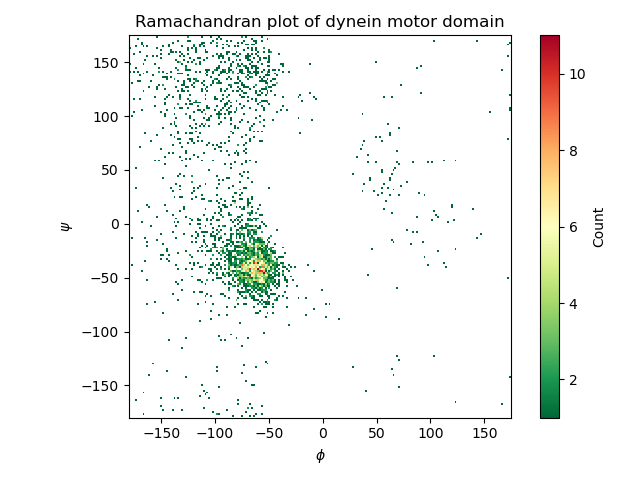
This script creates a Ramachandran plot of the motor domain of dynein (PDB: 3VKH). The protein mainly consists of alpha-helices, as the plot clearly indicates.
# Code source: Patrick Kunzmann
# License: BSD 3 clause
from tempfile import gettempdir
import matplotlib.pyplot as plt
import numpy as np
import biotite.database.rcsb as rcsb
import biotite.structure as struc
import biotite.structure.io as strucio
# Download and parse file
file = rcsb.fetch("3vkh", "cif", gettempdir())
atom_array = strucio.load_structure(file)
# Calculate backbone dihedral angles
# from one of the two identical chains in the asymmetric unit
phi, psi, omega = struc.dihedral_backbone(atom_array[atom_array.chain_id == "A"])
# Conversion from radians into degree
phi *= 180 / np.pi
psi *= 180 / np.pi
# Remove invalid values (NaN) at first and last position
phi = phi[1:-1]
psi = psi[1:-1]
# Plot density
figure = plt.figure()
ax = figure.add_subplot(111)
h, xed, yed, image = ax.hist2d(phi, psi, bins=(200, 200), cmap="RdYlGn_r", cmin=1)
cbar = figure.colorbar(image, orientation="vertical")
cbar.set_label("Count")
ax.set_aspect("equal")
ax.set_xlim(-180, 175)
ax.set_ylim(-180, 175)
ax.set_xlabel(r"$\phi$")
ax.set_ylabel(r"$\psi$")
ax.set_title("Ramachandran plot of dynein motor domain")
figure.tight_layout()
plt.show()