Note
Go to the end to download the full example code
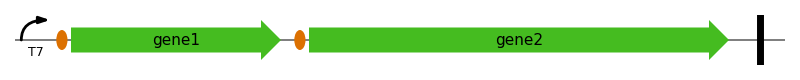
Feature map of a synthetic operon#
This script shows how to create a picture of an synthetic operon for publication purposes.
# Code source: Patrick Kunzmann
# License: BSD 3 clause
import matplotlib.pyplot as plt
import biotite.sequence.graphics as graphics
from biotite.sequence import Annotation, Feature, Location
strand = Location.Strand.FORWARD
prom = Feature(
"regulatory",
[Location(10, 50, strand)],
{"regulatory_class": "promoter", "note": "T7"},
)
rbs1 = Feature(
"regulatory",
[Location(60, 75, strand)],
{"regulatory_class": "ribosome_binding_site", "note": "RBS1"},
)
gene1 = Feature("gene", [Location(81, 380, strand)], {"gene": "gene1"})
rbs2 = Feature(
"regulatory",
[Location(400, 415, strand)],
{"regulatory_class": "ribosome_binding_site", "note": "RBS2"},
)
gene2 = Feature("gene", [Location(421, 1020, strand)], {"gene": "gene2"})
term = Feature(
"regulatory", [Location(1050, 1080, strand)], {"regulatory_class": "terminator"}
)
annotation = Annotation([prom, rbs1, gene1, rbs2, gene2, term])
fig = plt.figure(figsize=(8.0, 0.8))
ax = fig.add_subplot(111)
graphics.plot_feature_map(
ax,
annotation,
multi_line=False,
loc_range=(1, 1101),
)
fig.tight_layout()
plt.show()