Note
Go to the end to download the full example code
Cavity solvation in different states of an ion channel#
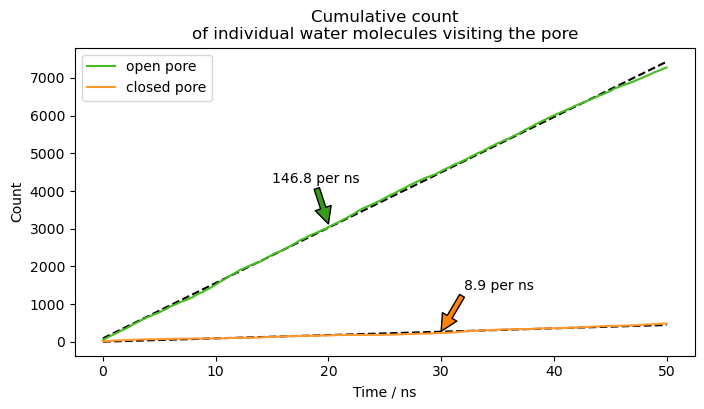
In selective sodium/potassium channels, the internal cavity of the pore is walled off from the solvent if the channel is closed. Upon activation, the internal gate opens and exchange of water molecules between the cavity and the bulk medium is possible.
Therefore, one can track the exchange rate of water molecules between the cavity and bulk to evaluate if a pore is open, closed, or in a transition between the two. Here, we used the distance between water molecules and residues located in the central cavity to evaluate if persistant water exchange takes place in different structures of the HCN4 channel.
The trajectories and template structure are not included in this example. However, the trajectories are based of publicly accessible structures of the open (PDB: 7NP3) and closed (PDB: 7NP4) state.
# Code source: Daniel Bauer, Patrick Kunzmann
# License: BSD 3 clause
import matplotlib.pyplot as plt
import numpy as np
from pylab import polyfit
import biotite
import biotite.structure as struct
import biotite.structure.io.gro as gro
import biotite.structure.io.xtc as xtc
def water_in_prox(atoms, sele, cutoff):
"""
Get the atom indices of water oxygen atoms that are in vicinity of
the selected atoms.
"""
cell_list = struct.CellList(atoms, cell_size=5, selection=atoms.atom_name == "OW")
adjacent_atoms = cell_list.get_atoms(atoms[sele].coord, cutoff)
adjacent_atoms = np.unique(adjacent_atoms.flatten())
adjacent_atoms = adjacent_atoms[adjacent_atoms > 0]
return adjacent_atoms
def cum_water_in_pore(traj, cutoff=6, key_residues=(507, 511)):
"""
Calculate the cumulative number of water molecules visiting the
pore.
"""
protein_sele = np.isin(traj.res_id, key_residues) & ~np.isin(
traj.atom_name, ["N", "O", "CA", "C"]
)
water_count = np.zeros(traj.shape[0])
prev_counted_indices = []
for idx, frame in enumerate(traj):
indices = water_in_prox(frame, protein_sele, cutoff)
count = (~np.isin(indices, prev_counted_indices)).sum()
if idx != 0:
count += water_count[idx - 1]
water_count[idx] = count
prev_counted_indices = indices
return water_count
# Calculate the cumulative number water molecules visiting the pore
# for the open and closed state
counts = []
for name in ["apo", "holo"]:
gro_file = gro.GROFile.read(f"{name}.gro")
template = gro_file.get_structure(model=1)
# Represent the water molecules by the oxygen atom
filter_indices = np.where(
struct.filter_amino_acids(template) | (template.atom_name == "OW")
)[0]
xtc_file = xtc.XTCFile.read(f"{name}.xtc", atom_i=filter_indices)
traj = xtc_file.get_structure(template[filter_indices])
cum_count = cum_water_in_pore(traj)
counts.append(cum_count)
time = np.arange(len(counts[0])) * 40 / 1000
# Linear fitting
open_fit = polyfit(time, counts[0], 1)
closed_fit = polyfit(time, counts[1], 1)
fig, ax = plt.subplots(figsize=(8.0, 4.0))
ax.plot(time, counts[0], label="open pore", color=biotite.colors["dimgreen"])
ax.plot(
time, open_fit[0] * time + open_fit[1], linestyle="--", color="black", zorder=-1
)
ax.plot(time, counts[1], label="closed pore", color=biotite.colors["lightorange"])
ax.plot(
time, closed_fit[0] * time + closed_fit[1], linestyle="--", color="black", zorder=-1
)
ax.set(
xlabel="Time / ns",
ylabel="Count",
title="Cumulative count\nof individual water molecules visiting the pore",
)
ax.legend()
ax.annotate(
f"{open_fit[0]:.1f} per ns",
xy=(20, 20 * open_fit[0] + open_fit[1] + 100),
xytext=(20 - 5, 20 * open_fit[0] + open_fit[1] + 1300),
arrowprops=dict(facecolor=biotite.colors["darkgreen"]),
va="center",
)
ax.annotate(
f"{closed_fit[0]:.1f} per ns",
xy=(30, 20 * closed_fit[0] + closed_fit[1] + 100),
xytext=(30 + 2, 20 * closed_fit[0] + closed_fit[1] + 1300),
arrowprops=dict(facecolor=biotite.colors["orange"]),
va="center",
)
fig.savefig("water_exchange.png", bbox_inches="tight")
plt.show()