Note
Go to the end to download the full example code
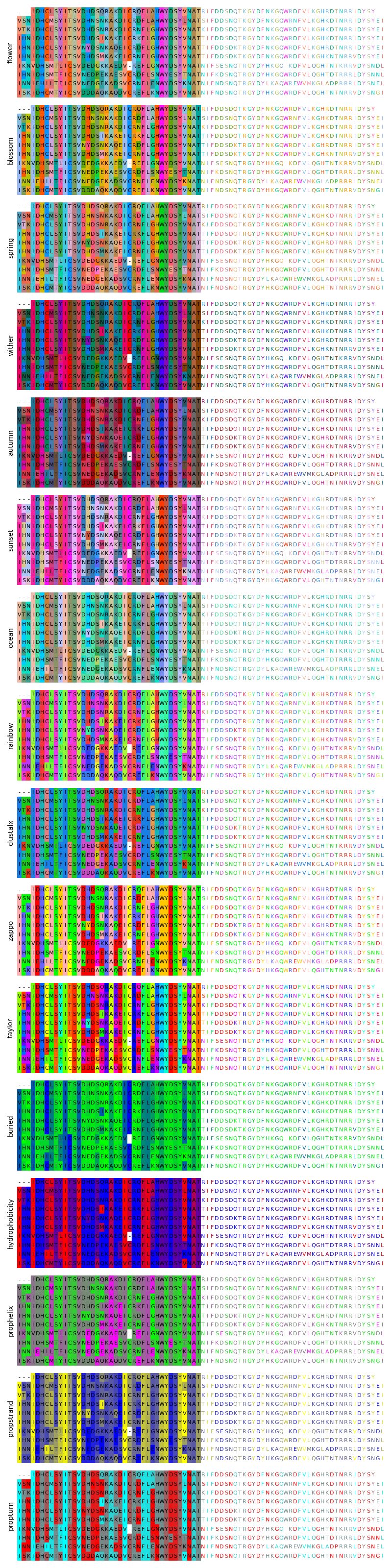
Biotite color schemes for protein sequences#
This script shows the same multiple protein sequence alignment in the different color schemes available in Biotite.
rainbow - Default color scheme in Biotite
clustalx - Default color scheme of the ClustalX software
Color schemes generated with the software Gecos [1]:
flower - Light color scheme, based on BLOSUM62
blossom - Light color scheme with high contrast, based on BLOSUM62, depicts symbol similarity worse than flower
spring - Light color scheme, based on BLOSUM62, with alanine fixed to gray
wither - Dark color scheme, analogous to blossom
autumn - Dark color scheme, analogous to spring
sunset - Red-green color vision deficiency adapated color scheme, based on BLOSUM62
ocean - Blue shifted, light color scheme, based on BLOSUM62
Color schemes adapted from JalView [2]:
zappo - Color scheme that depicts physicochemical properties
taylor - Color scheme invented by Willie Taylor
buried - Color scheme depicting the buried index
hydrophobicity - Color scheme depicting hydrophobicity
prophelix - Color scheme depicting secondary structure propensities
propstrand - Color scheme depicting secondary structure propensities
propturn - Color scheme depicting secondary structure propensities
# Code source: Patrick Kunzmann
# License: BSD 3 clause
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import biotite.database.entrez as entrez
import biotite.sequence as seq
import biotite.sequence.align as align
import biotite.sequence.graphics as graphics
import biotite.sequence.io.fasta as fasta
# Generate example alignment
# (the same as in the bacterial luciferase example)
query = entrez.SimpleQuery("luxA", "Gene Name") & entrez.SimpleQuery(
"srcdb_swiss-prot", "Properties"
)
uids = entrez.search(query, db_name="protein")
fasta_file = fasta.FastaFile.read(
entrez.fetch_single_file(uids, None, db_name="protein", ret_type="fasta")
)
sequences = [seq.ProteinSequence(seq_str) for seq_str in fasta_file.values()]
matrix = align.SubstitutionMatrix.std_protein_matrix()
alignment, order, _, _ = align.align_multiple(sequences, matrix)
# Order alignment according to the guide tree
alignment = alignment[:, order]
alignment = alignment[220:300]
# Get color scheme names
alphabet = seq.ProteinSequence.alphabet
schemes = [
"flower",
"blossom",
"spring",
"wither",
"autumn",
"sunset",
"ocean",
"rainbow",
"clustalx",
"zappo",
"taylor",
"buried",
"hydrophobicity",
"prophelix",
"propstrand",
"propturn",
]
count = len(schemes)
# Assert that this example displays all available amino acid color schemes
all_schemes = graphics.list_color_scheme_names(alphabet)
assert set(schemes) == set(all_schemes)
# Visualize each scheme using the example alignment
fig = plt.figure(figsize=(8.0, count * 2.0))
gridspec = GridSpec(2, count)
for i, name in enumerate(schemes):
for j, color_symbols in enumerate([False, True]):
ax = fig.add_subplot(count, 2, 2 * i + j + 1)
if j == 0:
ax.set_ylabel(name)
alignment_part = alignment[:40]
else:
alignment_part = alignment[40:]
graphics.plot_alignment_type_based(
ax,
alignment_part,
symbols_per_line=len(alignment_part),
color_scheme=name,
color_symbols=color_symbols,
symbol_size=8,
)
fig.tight_layout()
fig.subplots_adjust(wspace=0)
plt.show()