Note
Go to the end to download the full example code
Visualization of a custom plasmid#
This script shows how Feature objects are displayed in a
plasmid map by using a custom ‘toy’ Annotation.
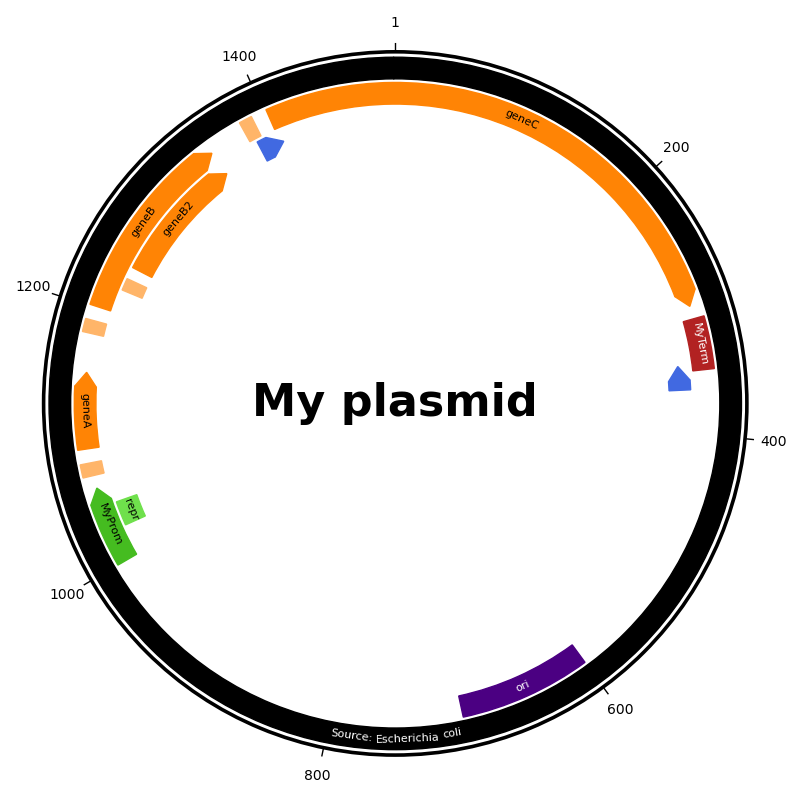
# Code source: Patrick Kunzmann
# License: BSD 3 clause
import matplotlib.pyplot as plt
import biotite.sequence as seq
import biotite.sequence.graphics as graphics
annotation = seq.Annotation(
[
seq.Feature(
"source", [seq.Location(0, 1500)], {"organism": "Escherichia coli"}
),
# Ori
seq.Feature(
"rep_origin",
[seq.Location(600, 700, seq.Location.Strand.REVERSE)],
{"regulatory_class": "promoter", "note": "MyProm"},
),
# Promoter
seq.Feature(
"regulatory",
[seq.Location(1000, 1060)],
{"regulatory_class": "promoter", "note": "MyProm"},
),
seq.Feature("protein_bind", [seq.Location(1025, 1045)], {"note": "repr"}),
# Gene A
seq.Feature(
"regulatory",
[seq.Location(1070, 1080)],
{"regulatory_class": "ribosome_binding_site"},
),
seq.Feature("CDS", [seq.Location(1091, 1150)], {"product": "geneA"}),
# Gene B
seq.Feature(
"regulatory",
[seq.Location(1180, 1190)],
{"regulatory_class": "ribosome_binding_site"},
),
seq.Feature("CDS", [seq.Location(1201, 1350)], {"product": "geneB"}),
seq.Feature(
"regulatory",
[seq.Location(1220, 1230)],
{"regulatory_class": "ribosome_binding_site"},
),
seq.Feature("CDS", [seq.Location(1240, 1350)], {"product": "geneB2"}),
# Gene C
seq.Feature(
"regulatory",
[seq.Location(1380, 1390)],
{"regulatory_class": "ribosome_binding_site"},
),
seq.Feature(
"CDS",
# CDS extends over periodic boundary -> two locations
[seq.Location(1, 300), seq.Location(1402, 1500)],
{"product": "geneC"},
),
# Terminator
seq.Feature(
"regulatory",
[seq.Location(310, 350)],
{"regulatory_class": "terminator", "note": "MyTerm"},
),
# Primers
# The labels will be too long to be displayed on the map
# If you want to display them nevertheless, set the
# 'omit_oversized_labels' to False
seq.Feature("primer_bind", [seq.Location(1385, 1405)], {"note": "geneC"}),
seq.Feature(
"primer_bind",
[seq.Location(345, 365, seq.Location.Strand.REVERSE)],
{"note": "geneC_R"},
),
# Terminator
seq.Feature(
"regulatory",
[seq.Location(310, 350)],
{"regulatory_class": "terminator", "note": "MyTerm"},
),
]
)
fig = plt.figure(figsize=(8.0, 8.0))
ax = fig.add_subplot(111, projection="polar")
graphics.plot_plasmid_map(
ax,
annotation,
plasmid_size=1500,
label="My plasmid",
label_properties={"fontsize": 8},
)
ticks = ax.get_xticks()
labels = ax.get_xticklabels()
fig.tight_layout()
plt.show()