Note
Go to the end to download the full example code
Customized visualization of a multiple sequence alignment#
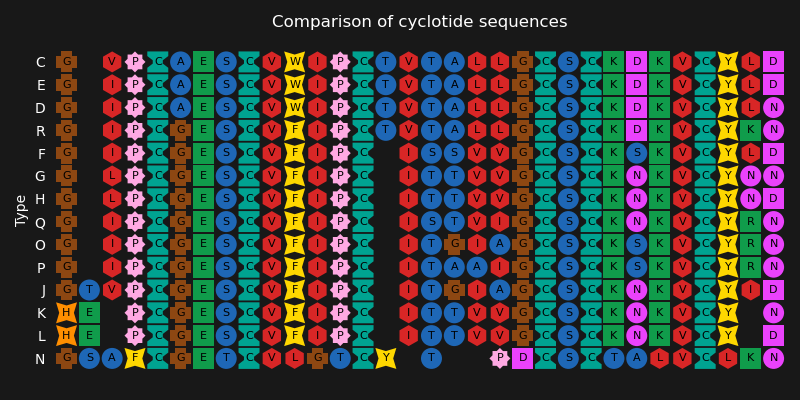
This example shows an alignment visualization used by the serious game Bionigma [1]. This visualization distinguishes amino acids by color and shape.
# Code source: Patrick Kunzmann
# License: BSD 3 clause
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.patches import Rectangle
from matplotlib.transforms import Bbox
import biotite.database.entrez as entrez
import biotite.sequence as seq
import biotite.sequence.align as align
import biotite.sequence.graphics as graphics
import biotite.sequence.io.fasta as fasta
# The polygon coordinates for the different shapes
_hexagon_coord = np.array(
[
(0.500, 0.000),
(0.937, 0.250),
(0.937, 0.750),
(0.500, 1.000),
(0.063, 0.750),
(0.063, 0.250),
]
)
_spiked_coord = np.array(
[
(0.000, 0.000),
(0.500, 0.150),
(1.000, 0.000),
(0.850, 0.500),
(1.000, 1.000),
(0.500, 0.850),
(0.000, 1.000),
(0.150, 0.500),
]
)
_spiked_coord = np.array(
[
(0.000, 0.000),
(0.500, 0.150),
(1.000, 0.000),
(0.850, 0.500),
(1.000, 1.000),
(0.500, 0.850),
(0.000, 1.000),
(0.150, 0.500),
]
)
_cross_coord = np.array(
[
(0.220, 0.000),
(0.780, 0.000),
(0.780, 0.220),
(1.000, 0.220),
(1.000, 0.780),
(0.780, 0.780),
(0.780, 1.000),
(0.220, 1.000),
(0.220, 0.780),
(0.000, 0.780),
(0.000, 0.220),
(0.220, 0.220),
]
)
_star_coord = np.array(
[
(0.500, 0.000),
(0.648, 0.150),
(0.852, 0.150),
(0.852, 0.352),
(1.000, 0.500),
(0.852, 0.648),
(0.852, 0.852),
(0.648, 0.852),
(0.500, 1.000),
(0.352, 0.852),
(0.148, 0.852),
(0.148, 0.648),
(0.000, 0.500),
(0.148, 0.352),
(0.148, 0.148),
(0.352, 0.148),
]
)
_hourglass_coord = np.array(
[
(0.000, 0.000),
(1.000, 0.000),
(1.000, 0.220),
(0.740, 0.420),
(0.740, 0.580),
(1.000, 0.780),
(1.000, 1.000),
(0.000, 1.000),
(0.000, 0.780),
(0.260, 0.580),
(0.260, 0.420),
(0.000, 0.220),
]
)
# The shape color for each symbols
_colors = {
"A": "#1e67b6",
"C": "#00a391",
"D": "#ea42fc",
"E": "#109c4b",
"F": "#fed700",
"G": "#8d4712",
"H": "#ff8e00",
"I": "#d82626",
"K": "#109c4b",
"L": "#d82626",
"M": "#d82626",
"N": "#ea42fc",
"P": "#ffa9e3",
"Q": "#109c4b",
"R": "#109c4b",
"S": "#1e67b6",
"T": "#1e67b6",
"V": "#d82626",
"W": "#fed700",
"Y": "#fed700",
}
class ShapePlotter(graphics.SymbolPlotter):
"""
A symbol plotter that depicts each symbol by color and shape.
"""
def __init__(self, axes, font_size=None, font_param=None):
super().__init__(axes)
# The symbol to shape mapping
self._draw_funcs = {
"A": ShapePlotter._draw_circle,
"T": ShapePlotter._draw_circle,
"S": ShapePlotter._draw_circle,
"N": ShapePlotter._draw_circle,
"D": ShapePlotter._draw_rectangle,
"E": ShapePlotter._draw_rectangle,
"Q": ShapePlotter._draw_rectangle,
"K": ShapePlotter._draw_rectangle,
"R": ShapePlotter._draw_rectangle,
"I": ShapePlotter._draw_hexagon,
"L": ShapePlotter._draw_hexagon,
"V": ShapePlotter._draw_hexagon,
"M": ShapePlotter._draw_hexagon,
"F": ShapePlotter._draw_spiked,
"W": ShapePlotter._draw_spiked,
"Y": ShapePlotter._draw_spiked,
"H": ShapePlotter._draw_spiked,
"G": ShapePlotter._draw_cross,
"P": ShapePlotter._draw_star,
"C": ShapePlotter._draw_hourglass,
}
self._font_size = font_size
self._font_param = font_param if font_param is not None else {}
def plot_symbol(self, bbox, alignment, column_i, seq_i):
trace = alignment.trace
if trace[column_i, seq_i] != -1:
symbol = alignment.sequences[seq_i][trace[column_i, seq_i]]
else:
symbol = ""
color = self._get_color(alignment, column_i, seq_i)
draw_func = self._draw_funcs.get(symbol)
# 'draw_func' is None for gaps
if draw_func is not None:
# Shrink Bbox slightly to get a small margin between shapes
f = 0.04
shape_bbox = Bbox(
(
(bbox.x0 + f * bbox.width, bbox.y0 + f * bbox.height),
(bbox.x1 - f * bbox.width, bbox.y1 - f * bbox.height),
),
)
draw_func(self, shape_bbox, color)
text = self.axes.text(
bbox.x0 + bbox.width / 2,
bbox.y0 + bbox.height / 2,
symbol,
color="black",
ha="center",
va="center",
size=self._font_size,
**self._font_param,
)
text.set_clip_on(True)
def _get_color(self, alignment, column_i, seq_i):
index = alignment.trace[column_i, seq_i]
if index == -1:
# Gaps are white
return (1, 1, 1)
code = alignment.sequences[seq_i].code[index]
return _colors[seq.ProteinSequence.alphabet.decode(code)]
def _draw_circle(self, bbox, color):
from matplotlib.patches import Circle
circle = Circle(
(bbox.x0 + bbox.width / 2, bbox.y0 + bbox.height / 2),
bbox.width / 2,
facecolor=color,
edgecolor="None",
fill=True,
)
self.axes.add_patch(circle)
def _draw_rectangle(self, bbox, color):
rectangle = Rectangle(
bbox.p0, bbox.width, bbox.height, facecolor=color, edgecolor="None"
)
self.axes.add_patch(rectangle)
def _draw_hexagon(self, bbox, color):
self._draw_polygon(bbox, color, _hexagon_coord)
def _draw_spiked(self, bbox, color):
self._draw_polygon(bbox, color, _spiked_coord)
def _draw_cross(self, bbox, color):
self._draw_polygon(bbox, color, _cross_coord)
def _draw_star(self, bbox, color):
self._draw_polygon(bbox, color, _star_coord)
def _draw_hourglass(self, bbox, color):
self._draw_polygon(bbox, color, _hourglass_coord)
def _draw_polygon(self, bbox, color, coord):
from matplotlib.patches import Polygon
# Transfom unit coordinates to fit Bbox
coord = coord.copy()
coord *= bbox.width
coord += bbox.p0
polygon = Polygon(coord, facecolor=color, edgecolor="None")
self.axes.add_patch(polygon)
def plot_alignment_shapes(
axes,
alignment,
symbols_per_line=30,
show_numbers=False,
number_size=None,
number_functions=None,
labels=None,
label_size=None,
show_line_position=False,
spacing=1,
symbol_size=None,
symbol_param=None,
):
"""
A thin wrapper around the 'ShapePlotter' and 'plot_alignment()'
function.
"""
symbol_plotter = ShapePlotter(axes, font_size=symbol_size, font_param=symbol_param)
graphics.plot_alignment(
axes=axes,
alignment=alignment,
symbol_plotter=symbol_plotter,
symbols_per_line=symbols_per_line,
show_numbers=show_numbers,
number_size=number_size,
number_functions=number_functions,
labels=labels,
label_size=label_size,
show_line_position=show_line_position,
spacing=spacing,
)
twin = axes.get_shared_x_axes().get_siblings(axes)[0]
for ax in (axes, twin):
ax.set_yticklabels(ax.get_yticklabels(), fontdict={"color": "white"})
axes.get_figure().patch.set_facecolor("#181818")
# Using cyclotide sequences as example
query = entrez.SimpleQuery("Cyclotide") & entrez.SimpleQuery(
"cter"
) & entrez.SimpleQuery("srcdb_swiss-prot", field="Properties") ^ entrez.SimpleQuery(
"Precursor"
)
uids = entrez.search(query, "protein")
fasta_file = fasta.FastaFile.read(
entrez.fetch_single_file(uids, None, "protein", "fasta")
)
sequence_dict = fasta.get_sequences(fasta_file)
# Currently there seems to b a bug in the NCBI search,
# so that 'Precursor' results are still included
# Solve this by filtering the sequence length
sequence_dict = {header: seq for header, seq in sequence_dict.items() if len(seq) < 100}
headers = list(sequence_dict.keys())
sequences = list(sequence_dict.values())
labels = [header[-1] for header in headers]
# Perform a multiple sequence alignment
matrix = align.SubstitutionMatrix.std_protein_matrix()
alignment, order, _, _ = align.align_multiple(sequences, matrix)
# Order alignment according to guide tree
alignment = alignment[:, order.tolist()]
labels = [labels[i] for i in order]
# Visualize the alignment using the new alignment plotter
fig = plt.figure(figsize=(8.0, 4.0))
ax = fig.add_subplot(111)
plot_alignment_shapes(
ax, alignment, labels=labels, symbols_per_line=len(alignment), symbol_size=8
)
# The aspect ratio of the shapes should be preserved:
# Squares should look like squares, circles should look like circles
ax.set_aspect("equal")
ax.set_ylabel("Type", color="white")
ax.set_title("Comparison of cyclotide sequences", color="white")
fig.tight_layout()
plt.show()