Note
Go to the end to download the full example code
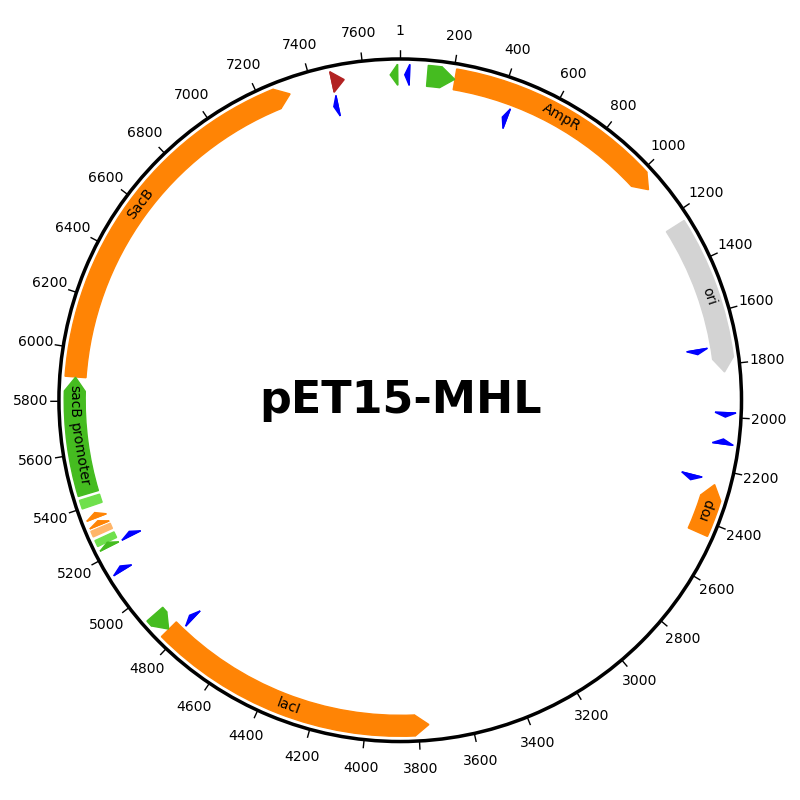
Plasmid map of a vector#
This script downloads the GenBank file for a pET15 plasmid from AddGene and draws a plasmid map using a custom feature formatter.
Promoters - green arrow
Terminators - red arrow
Protein binding sites - light green rectangle
RBS - light orange rectangle
CDS - orange arrow
Ori - gray arrow
Primer - blue arrow
# Code source: Patrick Kunzmann
# License: BSD 3 clause
import io
import matplotlib.pyplot as plt
import requests
import biotite
import biotite.sequence.graphics as graphics
import biotite.sequence.io.genbank as gb
PLASMID_URL = (
"https://media.addgene.org/snapgene-media/v1.7.9-0-g88a3305/"
"sequences/12250/9998fdbe-051f-4dc6-ba0f-24e65127a0c5/"
"addgene-plasmid-26092-sequence-12250.gbk"
)
response = requests.get(PLASMID_URL)
gb_file = gb.GenBankFile.read(io.StringIO(response.text))
annotation = gb.get_annotation(
gb_file,
include_only=[
"promoter",
"terminator",
"protein_bind",
"RBS",
"CDS",
"rep_origin",
"primer_bind",
],
)
_, seq_length, _, _, _, _ = gb.get_locus(gb_file)
# AddGene stores the plasmid name in the 'KEYWORDS' field
# [0][0][0] ->
# The first (and only) 'KEYWORDS' field
# The first entry in the tuple
# The first (and only) line in the field
plasmid_name = gb_file.get_fields("KEYWORDS")[0][0][0]
def custom_feature_formatter(feature):
# AddGene stores the feature label in the '\label' qualifier
label = feature.qual.get("label")
if feature.key == "promoter":
return True, biotite.colors["dimgreen"], "black", label
elif feature.key == "terminator":
return True, "firebrick", "black", label
elif feature.key == "protein_bind":
return False, biotite.colors["lightgreen"], "black", label
elif feature.key == "RBS":
return False, biotite.colors["brightorange"], "black", label
elif feature.key == "CDS":
return True, biotite.colors["orange"], "black", label
elif feature.key == "rep_origin":
return True, "lightgray", "black", label
elif feature.key == "primer_bind":
return True, "blue", "black", label
fig = plt.figure(figsize=(8.0, 8.0))
ax = fig.add_subplot(111, projection="polar")
graphics.plot_plasmid_map(
ax,
annotation,
plasmid_size=seq_length,
label=plasmid_name,
feature_formatter=custom_feature_formatter,
)
fig.tight_layout()
plt.show()