Note
Go to the end to download the full example code
Biotite color schemes#
This script displays the available color schemes.
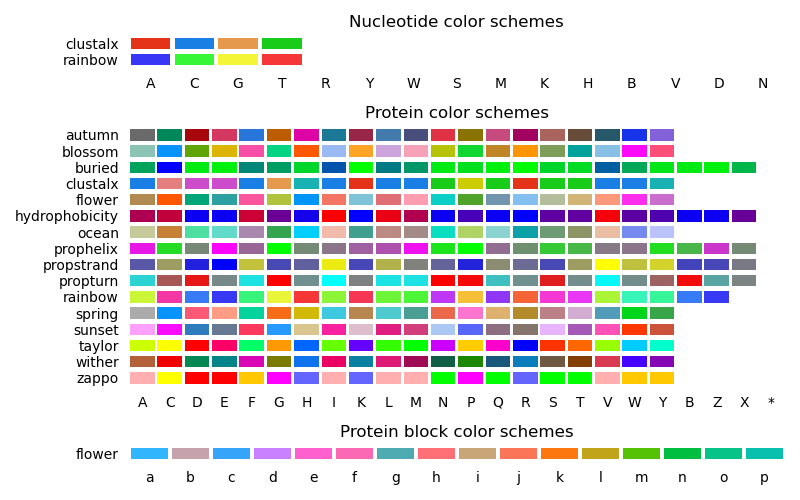
# Code source: Patrick Kunzmann
# License: BSD 3 clause
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.gridspec import GridSpec
from matplotlib.patches import Rectangle
import biotite.sequence as seq
import biotite.sequence.graphics as graphics
def plot_colors(ax, alphabet):
x_space = 0.1
y_space = 0.3
scheme_names = sorted(graphics.list_color_scheme_names(alphabet))
scheme_names.reverse()
schemes = [graphics.get_color_scheme(name, alphabet) for name in scheme_names]
for i, scheme in enumerate(schemes):
for j, color in enumerate(scheme):
box = Rectangle(
(j - 0.5 + x_space / 2, i - 0.5 + y_space / 2),
1 - x_space,
1 - y_space,
color=color,
linewidth=0,
)
ax.add_patch(box)
ax.set_xticks(np.arange(len(alphabet)))
ax.set_yticks(np.arange(len(schemes)))
ax.set_xticklabels([symbol for symbol in alphabet])
ax.set_yticklabels(scheme_names)
ax.set_xlim(-0.5, len(alphabet) - 0.5)
ax.set_ylim(-0.5, len(schemes) - 0.5)
ax.spines["left"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.spines["bottom"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.xaxis.set_ticks_position("none")
ax.yaxis.set_ticks_position("none")
nuc_alphabet = seq.NucleotideSequence.alphabet_amb
prot_alphabet = seq.ProteinSequence.alphabet
pb_alphabet = seq.LetterAlphabet("abcdefghijklmnop")
figure = plt.figure(figsize=(8.0, 5.0))
gs = GridSpec(
3,
1,
height_ratios=[
len(graphics.list_color_scheme_names(alphabet))
for alphabet in (nuc_alphabet, prot_alphabet, pb_alphabet)
],
)
ax = figure.add_subplot(gs[0, 0])
ax.set_title("Nucleotide color schemes")
plot_colors(ax, nuc_alphabet)
ax = figure.add_subplot(gs[1, 0])
ax.set_title("Protein color schemes")
plot_colors(ax, prot_alphabet)
ax = figure.add_subplot(gs[2, 0])
ax.set_title("Protein block color schemes")
plot_colors(ax, pb_alphabet)
plt.tight_layout()
plt.show()