Note
Go to the end to download the full example code
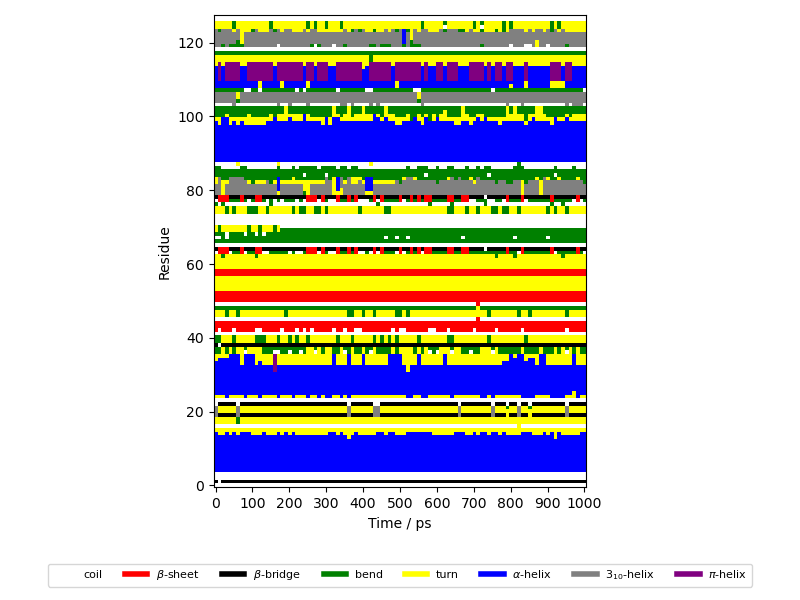
Secondary structure during an MD simulation#
This script displays positional changes of secondary structure elements (SSE) in the course of time of an MD simulation.
The trajectory file can be downloaded
here
and the template PDB can be downloaded
here.
# Code source: Daniel Bauer, Patrick Kunzmann
# License: BSD 3 clause
import matplotlib.pyplot as plt
import numpy as np
from matplotlib import colors
from matplotlib.lines import Line2D
import biotite.structure as struc
import biotite.structure.io as strucio
import biotite.structure.io.xtc as xtc
from biotite.application.dssp import DsspApp
# Put here the path of the downloaded files
templ_file_path = "../../../download/lysozyme_md.pdb"
traj_file_path = "../../../download/lysozyme_md.xtc"
xtc_file = xtc.XTCFile.read(traj_file_path)
traj = xtc_file.get_structure(template=strucio.load_structure(templ_file_path))
time = xtc_file.get_time()
traj = traj[:, struc.filter_amino_acids(traj)]
# DSSP does not assign an SSE to the last residue -> -1
sse = np.empty((traj.shape[0], struc.get_residue_count(traj) - 1), dtype="U1")
for idx, frame in enumerate(traj):
app = DsspApp(traj[idx])
app.start()
app.join()
sse[idx] = app.get_sse()
# Matplotlib needs numbers to assign colors correctly
def sse_to_num(sse):
num = np.empty(sse.shape, dtype=int)
num[sse == "C"] = 0
num[sse == "E"] = 1
num[sse == "B"] = 2
num[sse == "S"] = 3
num[sse == "T"] = 4
num[sse == "H"] = 5
num[sse == "G"] = 6
num[sse == "I"] = 7
return num
sse = sse_to_num(sse)
# Plotting
# SSE colormap
color_assign = {
r"coil": "white",
r"$\beta$-sheet": "red",
r"$\beta$-bridge": "black",
r"bend": "green",
r"turn": "yellow",
r"$\alpha$-helix": "blue",
r"$3_{10}$-helix": "gray",
r"$\pi$-helix": "purple",
}
cmap = colors.ListedColormap(color_assign.values())
plt.figure(figsize=(8.0, 6.0))
plt.imshow(sse.T, cmap=cmap, origin="lower")
plt.xlabel("Time / ps")
plt.ylabel("Residue")
ticks = np.arange(0, len(traj), 10)
plt.xticks(ticks, time[ticks].astype(int))
# Custom legend below the DSSP plot
custom_lines = [Line2D([0], [0], color=cmap(i), lw=4) for i in range(len(color_assign))]
plt.legend(
custom_lines,
color_assign.keys(),
loc="upper center",
bbox_to_anchor=(0.5, -0.15),
ncol=len(color_assign),
fontsize=8,
)
plt.tight_layout()
plt.show()